How to Read 5 Line of Text File Using Pandas
What is a fixed width text file?
A fixed width file is similar to a csv file, simply rather than using a delimiter, each field has a set number of characters. This creates files with all the data tidily lined upward with an appearance similar to a spreadsheet when opened in a text editor. This is convenient if you're looking at raw data files in a text editor, merely less ideal when you need to programmatically work with the data.
Fixed width files accept a few common quirks to keep in mind:
- When values don't consume the total graphic symbol count for a field, a padding graphic symbol is used to bring the character count up to the total for that field.
- Whatever character can be used equally a padding character as long equally it is consequent throughout the file. White space is a common padding character.
- Values tin exist left or correct aligned in a field and alignment must exist consistent for all fields in the file.
A thorough description of a fixed width file is available here.
Note : All fields in a fixed width file practice non need to have the same character count. For example: in a file with three fields, the offset field could be 6 characters, the second 20, and the concluding nine.
How to spot a fixed width text file?
Upon initial examination, a fixed width file tin can look like a tab separated file when white space is used as the padding character. If you're trying to read a fixed width file as a csv or tsv and getting mangled results, try opening it in a text editor. If the information all line up tidily, it'due south probably a stock-still width file. Many text editors also requite graphic symbol counts for cursor placement, which makes information technology easier to spot a pattern in the character counts.
If your file is as well large to hands open in a text editor, there are diverse ways to sample portions of it into a separate, smaller file on the command line. An easy method on a Unix/Linux arrangement is the head command. The example below uses head with -n 50 to read the commencement 50 lines of large_file.txt so copy them into a new file called first_50_rows.txt.
head -n l large_file.txt > first_50_rows.txt Allow's work with a real life instance file
UniProtKB Database
The UniProt Knowledgebase (UniProtKB) is a freely accessible and comprehensive database for protein sequence and note data bachelor nether a CC-BY (4.0) license. The Swiss-Prot co-operative of the UniProtKB has manually annotated and reviewed information well-nigh proteins for various organisms. Consummate datasets from UniProt information tin be downloaded from ftp.uniprot.org. The data for human proteins are contained in a prepare of stock-still width text files: humchr01.txt - humchr22.txt, humchrx.txt, and humchry.txt.
We don't need all 24 files for this example, so here's the link to the showtime file in the fix:
https://ftp.uniprot.org/pub/databases/uniprot/current_release/knowledgebase/complete/docs/humchr01.txt
Examine the file before reading it with pandas
A quick glance at the file in a text editor shows a substantial header that we don't demand leading into 6 fields of data.
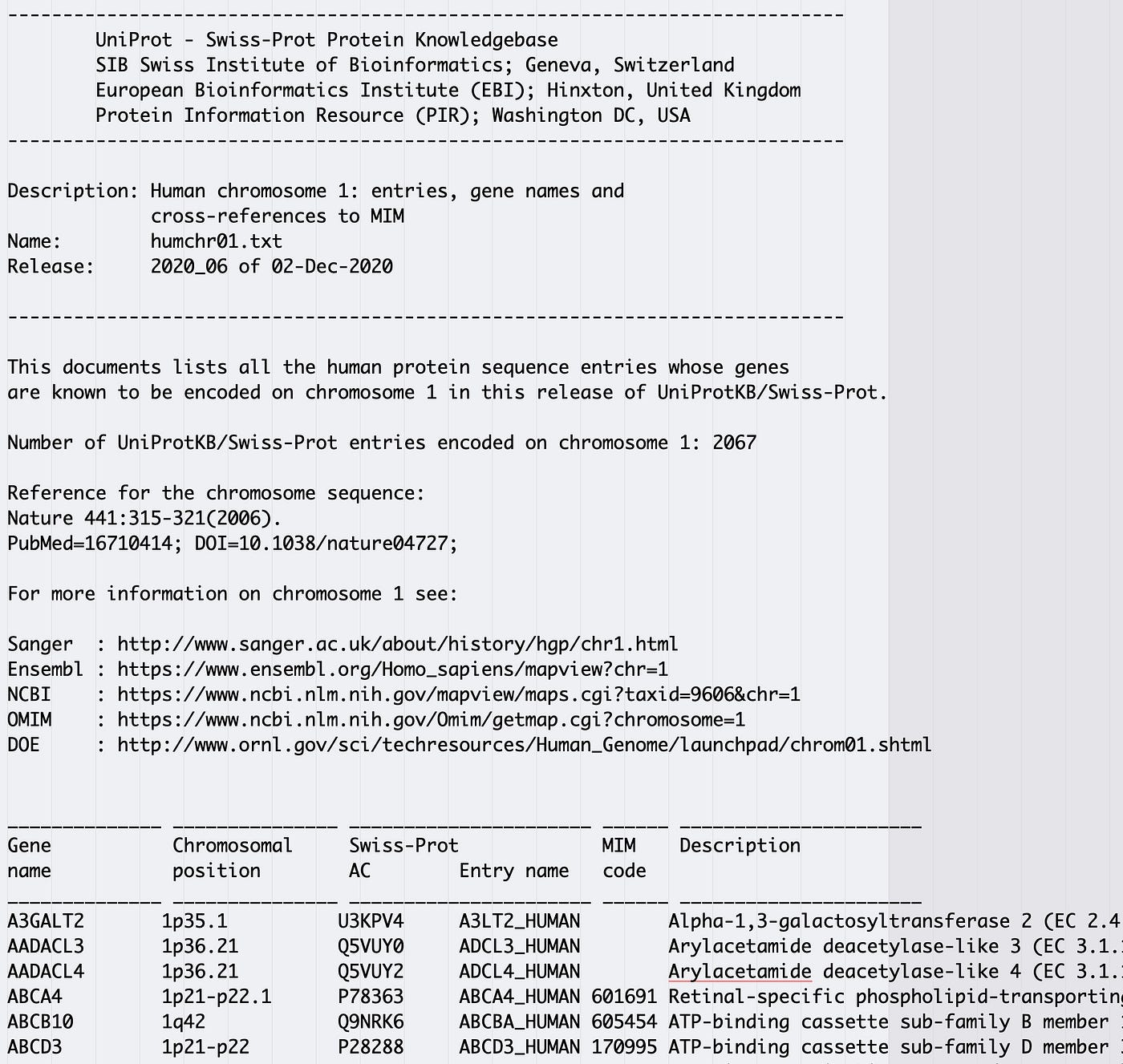
Stock-still width files don't seem to be as common as many other data file formats and they can look similar tab separated files at first glance. Visual inspection of a text file in a good text editor before trying to read a file with Pandas tin substantially reduce frustration and assist highlight formatting patterns.
Using pandas.read_fwf() with default parameters
Note: All lawmaking for this example was written for Python3.6 and Pandas1.2.0.
The documentation for pandas.read_fwf() lists 5 parameters:
filepath_or_buffer, colspecs, widths, infer_nrows, and **kwds
Two of the pandas.read_fwf() parameters, colspecs and infer_nrows, have default values that piece of work to infer the columns based on a sampling of initial rows.
Let's apply the default settings for pandas.read_fwf() to get our tidy DataFame. Nosotros'll leave the colspecs parameter to its default value of 'infer', which in turn utilizes the default value (100) of the infer_nrows parameter. These two defaults attempt to find a blueprint in the first 100 rows of data (subsequently any skipped rows) and apply that pattern to separate the data into columns.
Bones file cleanup
In that location are several rows of file header that precede the tabular info in our example file. We demand to skip them when we read the file.
None of the parameters seem ideal for skipping rows when reading the file. So how do we exercise it? We utilize the **kwds parameter.
Conveniently, pandas.read_fwf() uses the same TextFileReader context manager equally pandas.read_table(). This combined with the **kwds parameter allows us to use parameters for pandas.read_table() with pandas.read_fwf(). So we tin use the skiprows parameter to skip the first 35 rows in the example file. Similarly, we can use the skipfooter parameter to skip the terminal five rows of the example file that contain a footer that isn't role of the tabular data.
pandas.read_fwf('humchr01.txt', skiprows=35, skipfooter=five) The above endeavor leaves the DataFrame a bit of a mess 😔:
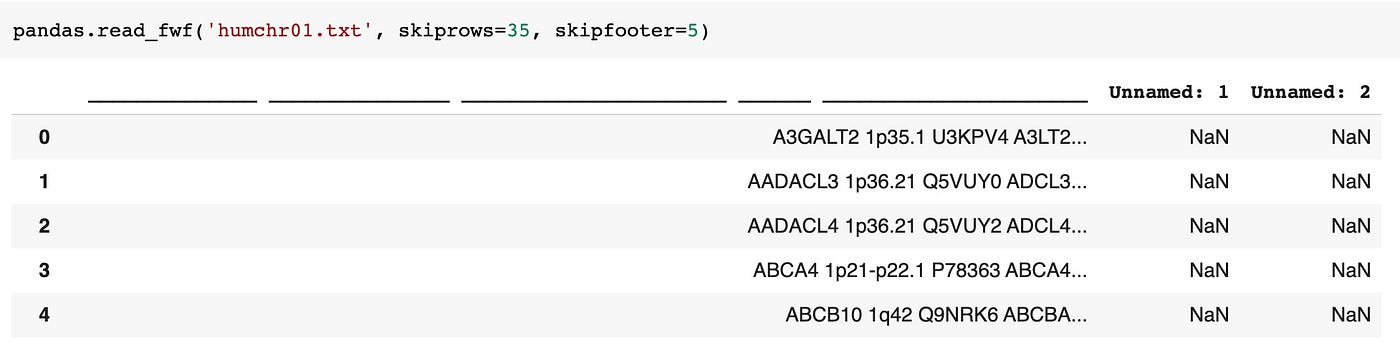
Annotation: Since we're using the default values for colspecs and infer_nrows we don't have to declare them.
Part of the upshot here is that the default colspecs parameter is trying to infer the column widths based on the showtime 100 rows, but the row right earlier the tabular data (row 36 in the file and shown in the column names above) doesn't really follow the graphic symbol count patterns in the data table, so the inferred cavalcade widths are getting mangled.
If nosotros'd set skiprows to 36 instead of 35, we'd have concluded upwardly with the first row of data pushed into the column names, which as well mangles the inferred column widths. At that place's no winning hither without some additional cleanup. Let's settle the cavalcade names issue with the names parameter and see if that helps.
Notation: Using the names parameter means we are not allocating a row in the file to cavalcade names, and then we every bit users have to brand sure to business relationship for the fact that skiprows must start at the first data row. So skiprows is set to 36 in the next instance but it was 35 in previous examples when we didn't utilise the names parameter.
pandas.read_fwf('humchr01.txt', skiprows=36, skipfooter=5, names=['gene_name', 'chromosomal_position', 'uniprot', 'entry_name', 'mtm_code', 'description']) 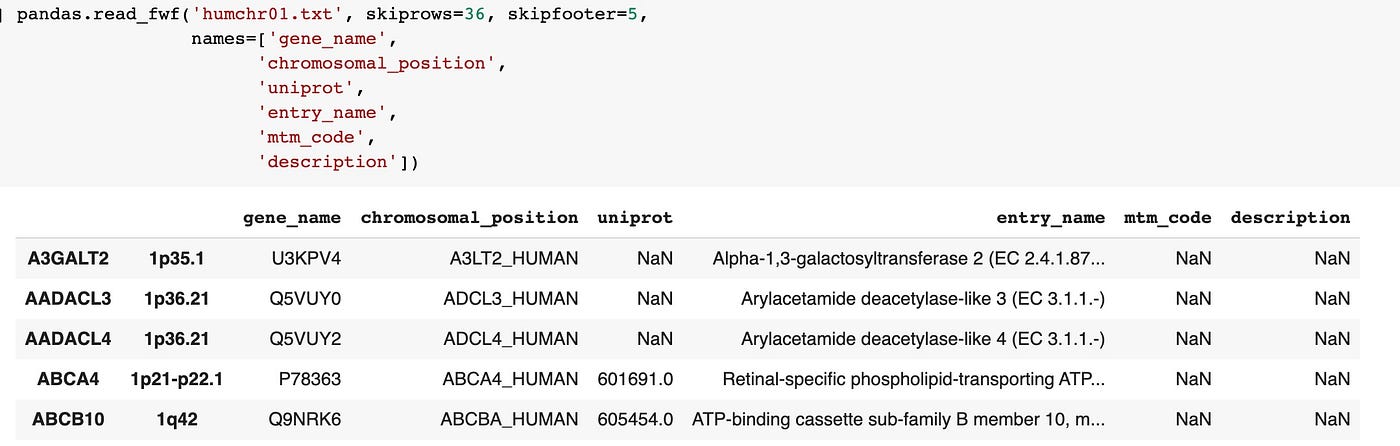
That's meliorate, but still a bit of a mess. Pandas inferred the cavalcade splits correctly, simply pushed the first 2 fields to the index. Let'due south fix the alphabetize issue past setting index_col=Imitation.
pandas.read_fwf('humchr01.txt', skiprows=36, skipfooter=5, index_col=False, names=['gene_name', 'chromosomal_position', 'uniprot', 'entry_name', 'mtm_code', 'description']) 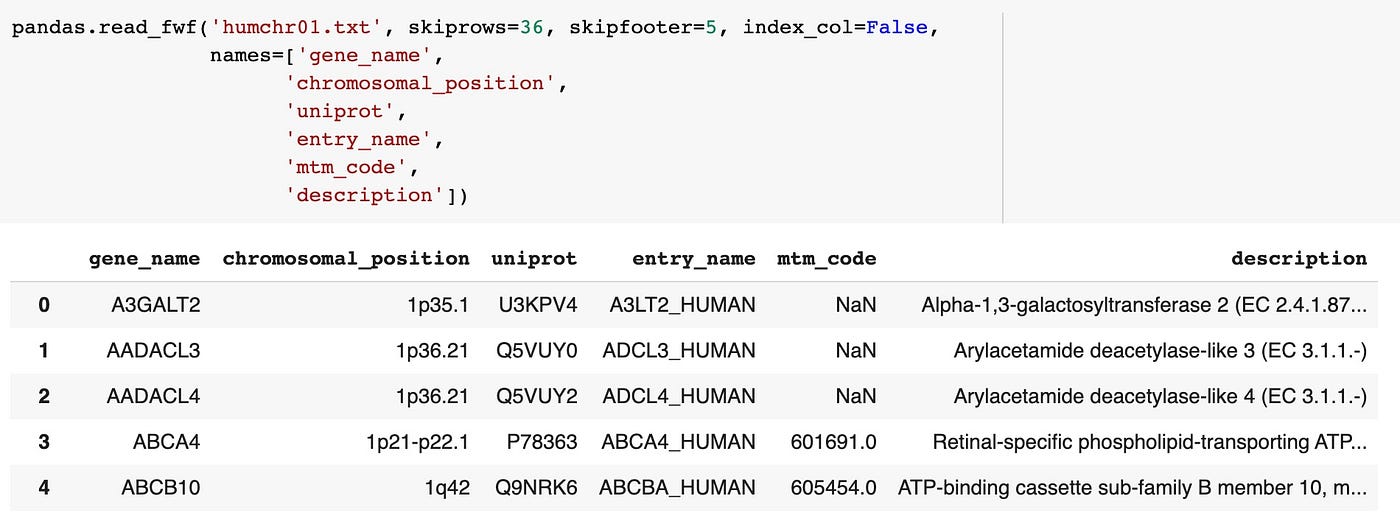
That looks good! The columns are split correctly, the column names make sense and the beginning row of data in the DataFrame matches the first row in the example file.
We relied on the default settings for two of the pandas.read_fwf() specific parameters to get our tidy DataFame. The colspecs parameter was left to its default value of 'infer' which in turn utilizes the default value of the infer_nrows parameter and finds a blueprint in the starting time 100 rows of data (subsequently the skipped rows) and uses that to split the data into columns. The default parameters worked well for this example file, simply nosotros could likewise specify the colspecs parameter instead of letting pandas infer the columns.
Setting field widths manually with colspecs
Simply like with the example higher up, we need to commencement with some bones cleanup. We'll drib the header and footer in the file and set the column names just like before.
The side by side pace is to build a list of tuples with the intervals of each field. The list beneath fits the example file.
colspecs = [(0, 14), (14, 30), (xxx, 41), (41, 53), (53, 60), (60, -1)] Note the terminal tuple: (60, -1). We can apply -ane to indicate the last index value. Alternately, we could use None instead of -i to indicate the last index value.
Note: When using colspecs the tuples don't take to be exclusionary! The last columns tin can exist set to tuples that overlap if that is desired. For example, if you want the first field duplicated: colspecs = [(0, 14), (0, 14), ...
pandas.read_fwf('humchr01.txt', skiprows=36, skipfooter=5, colspecs=colspecs, names=['gene_name', 'chromosomal_position', 'uniprot', 'entry_name', 'mtm_code', 'description']) 
Once more than we've attained a tidy DataFrame. This time we explicitly declared our field start and finish positions using the colspecs parameter rather than letting pandas infer the fields.
Conclusion
Reading fixed width text files with Pandas is easy and attainable. The default parameters for pandas.read_fwf() piece of work in nearly cases and the customization options are well documented. The Pandas library has many functions to read a diverseness of file types and the pandas.read_fwf() is one more useful Pandas tool to keep in mind.
courticechand1991.blogspot.com
Source: https://towardsdatascience.com/parsing-fixed-width-text-files-with-pandas-f1db8f737276
0 Response to "How to Read 5 Line of Text File Using Pandas"
Post a Comment